DNA analysis is considered the gold standard of forensic sciences because the scientific basis for profile comparison is well grounded, the error rates are quantifiable and usually very small, and the development procedure is highly reliable.
More goes into DNA analysis than the outsider might think. Forensic DNA analysts do not develop a full genomic profile (which would take a substantial amount of time and resources); rather, they look at very small parts of the genome where there is high variability among the population.
There are several forms of DNA analysis, but the most common one, STR, or short tandem repeat, examines the repetitions of base sequences at several locations of non-coding, nuclear DNA. Non-coding refers to the fact that there are locations on the DNA which do not code for specific protein sequences or functions, popularly referred to as “junk DNA”. We inherit one set of chromosomes from each parent and so have two alleles at each location (or locus).
What does a DNA report look like?
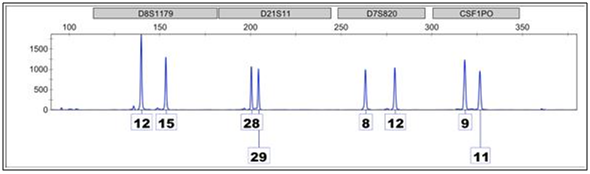
This is an electropherogram of four loci, represented by the grey boxes at the top, give the scientific nomenclature associated with that location on the genome.
The peaks signify alleles, or versions of that location, which are present in the sample. The number represents how many times a short sequence of nucleotide is repeated at a particular location on the genome in each version. The individual who contributed to this sample, for example, is a(12, 15) at D8, a (28, 29) at D2, a (8, 12) at D7, and a (9, 11) at CSF. This electropherogram is an example of a clean, single-source profile.
At D8, one parent contributed a chromosome with short sequence repeated 12 times, while the other parent contributed a chromosome where the same short sequence repeated 15 times. If an individual inherits the same allele from each parent, they are homozygous at that locus.
If the profile that the lab develops from evidence at the scene matches the profile of the defendant, a DNA expert will likely come to court to explain the analysis process and declare what the strength of the DNA evidence is. Usually, a statistic known as the random match probability describes the strength of the match between the two profiles.
The random match probability (RMP) reflects the profile’s rarity and gives us the likelihood that a randomly chosen, unrelated individual from a given population would have the same DNA profile observed in a sample. It relies on several key assumptions and, in some cases, can be controversial. In cases where the suspect has come under suspicion because of non-DNA evidence, and the DNA profile developed from the crime scene is a single-source full profile, most statisticians agree that the RMP is the most accurate measure of a coincidental match.
DNA, like other trace evidence, is only sometimes discovered and can be mixed, degraded, or contaminated. Even when DNA has been collected and properly stored and analyzed, the end result may only be a partial profile. Partial profiles don’t show alleles at all thirteen locations, but may only show alleles at varied locations, making the statistic associated less meaningful.
If there is a very small amount of DNA, interpretation of the results becomes considerably more complicated. Throughout analysis, the interpretation of artifacts like blobs, stutters, and off-ladder peaks can mean the difference between a match and an inconclusive result, or an inconclusive result and an exclusion. This is why labs are required to validate each piece of equipment and assess what levels of DNA input can still yield reliable results and are too low to be analyzed “below threshold”.
DNA Mixtures
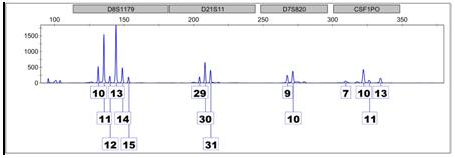
When more than one individual has contributed DNA to a sample, the result is a mixture. The electropherogram above is an example because there are more than two peaks at multiple locations. Looking at D8 – the first locus on the left – we see six peaks of varying heights, indicating at least, or perhaps more than, three different contributors. Differences in peak height can be attributed to homozygous contributors or processes during the amplification phases of analysis.
How is a random match probability calculated?
If the sample comes from a single-source profile, calculating the random match probability is straightforward. The RMP relies on what is called the product rule: the probability of multiple independent events occurring is the product of the probabilities of each event occurring.
For example, the probability of getting two heads in a row on the flip of a fair coin is 0.5 * 0.5 or 0.25. There is a 50% chance that we get heads on the first flip and a 50% chance that we get heads on the second, thus a 25% chance that we would get two heads in a row.
In DNA analysis, each locus present qualifies as an independent event since the loci chosen have satisfied certain tests indicating that which alleles inherited at one location does not impact alleles inherited at another location. The frequency of each allele in the population can be used to calculate the frequency of the genotype at each locus, all of which can then be multiplied together. When some of the alleles are not present or if there are multiple contributors to the sample, calculating a statistic like the RMP becomes considerably challenging.
One of the costs of performing DNA analysis is the consumption of the biological material tested. In cases where only a small amount of DNA was collected, consuming all of the DNA evidence means that it can’t be retested. If a mistake occurs during testing (contamination, artifacts, etc.), it can only be reinterpreted. Forensic laboratories should consume no more than is necessary to test in order to preserve the possibility of a retest, as retests have prevented many miscarriages of justice.
A great deal of information about forensic STR typing can be found here.